Feature#
Motivition
Computational analysis and modeling of transmembrane protein sequences and structures involve a complex workflow that requires integrating multiple tools for sequence alignment, topology prediction, structural analysis, and functional annotation. These tasks often demand extensive manual effort and switching between different software environments, making the process time-consuming and prone to inconsistencies.
What has been done
For this reason, we developed TMKit to offer a comprehensive and unified computational framework. It simplifies and standardises transmembrane protein analysis. TMKit consolidates various computational procedures under a single, intuitive interface, enabling researchers to efficiently perform sequence retrieval, topology parsing, structure comparison, and contact prediction without the need for multiple disparate tools.
Result
By reducing complexity and automating key steps, TMKit significantly lowers the computational workload, enhances data consistency, and facilitates seamless transmembrane protein research from sequence to structure.
TMKit provides currently nine modules to handle a number of transmembrane protein sequence and structural analysis problems, including visualization, sequence, quality control, topology, mapping, annotation, connectivity, edge extraction, and feature.
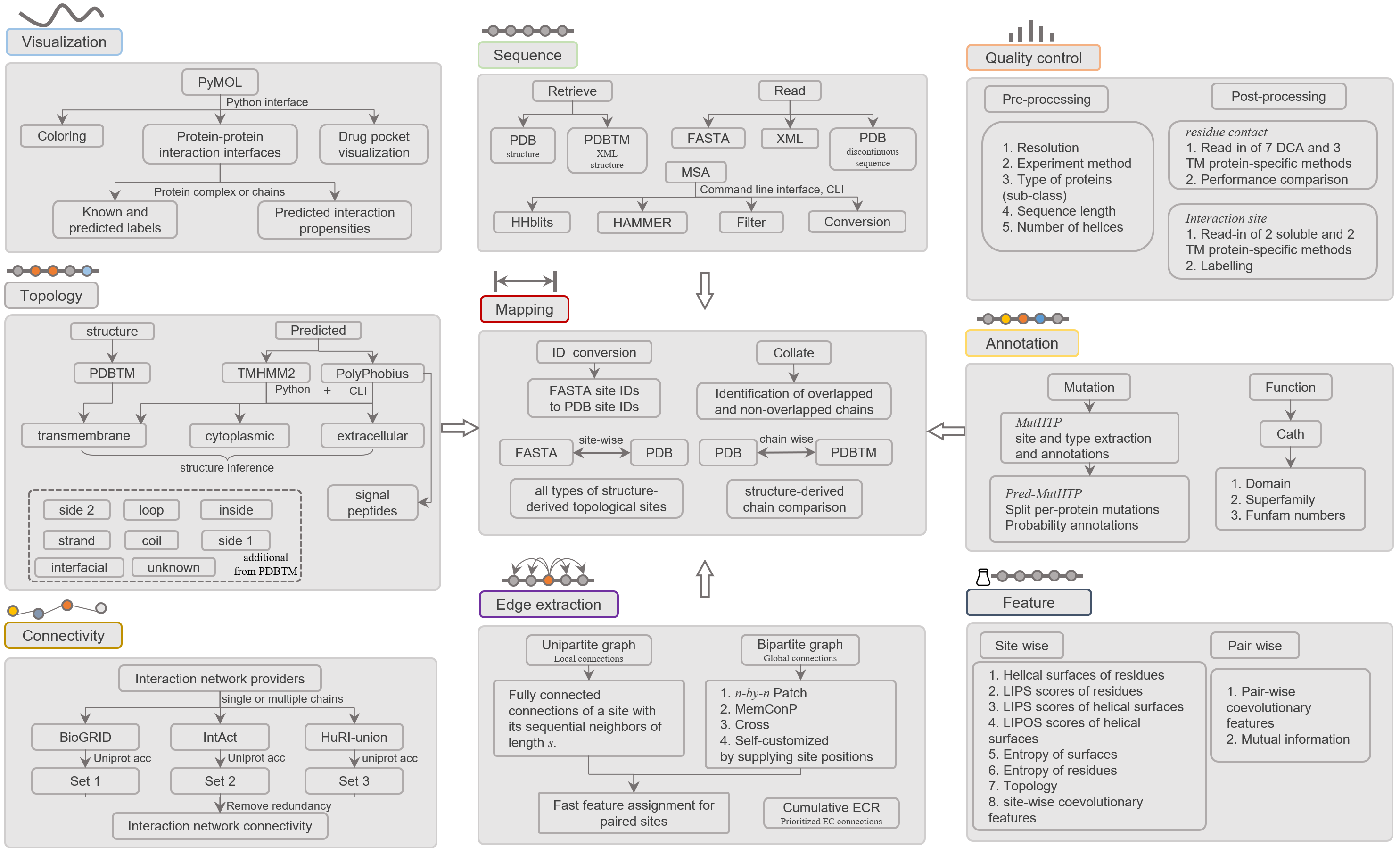
Caption: Functionality of TMKit.#
Tip
Visualization, Mapping, Annotation, Connectivity, and Edge extraction modules in TMKit are considered unique compared to existing tools.
