Coloring#
TMKit allows users to color a protein in its any segments. This function is added to TMKit considering that users may simply want to highlight some important domains and see clearly where they appear in a protein structure. This is exactly when you can consider this application. We made it readily available by using tmk.vs.coloring.
Reminder of data
Please make sure that the build-in example dataset has been downloaded before you walk through the tutorial.
Example usage#
As an example, we take protein 6uiw chain A to color the following segments, residues 1-4, 58-61, 5-57, 62-81, 62, 78, and 81. A segment you want to color can be a single residue or a list of continuously numbered residues. First, the whole protein will be colored chromium. Then, if you want to color residues 1-4 red, residues 5-57 orange, residues 58-61 red, residues 62-81 br4, and particularly, residues 62, 78, and 81 violet, you can put parameters actions and colors that way as shown below. The forms parameter in the command will render the segments in lines form.
import tmkit as tmk
tmk.vs.coloring(
pdb_fp='data/pdb/',
prot_name='6uiw',
seq_chain='A',
prot_c='chromium',
names=['segment1', 'segment2', 'segment3', 'segment4', 'segment5'],
actions=['resi 1-4', 'resi 5-57', 'resi 58-61', 'i. 62-81', 'i. 62+78+81'],
colors=['red', 'orange', 'red', 'br4', 'violet', ],
forms=['lines', 'lines', 'lines', 'lines', 'lines', ],
)
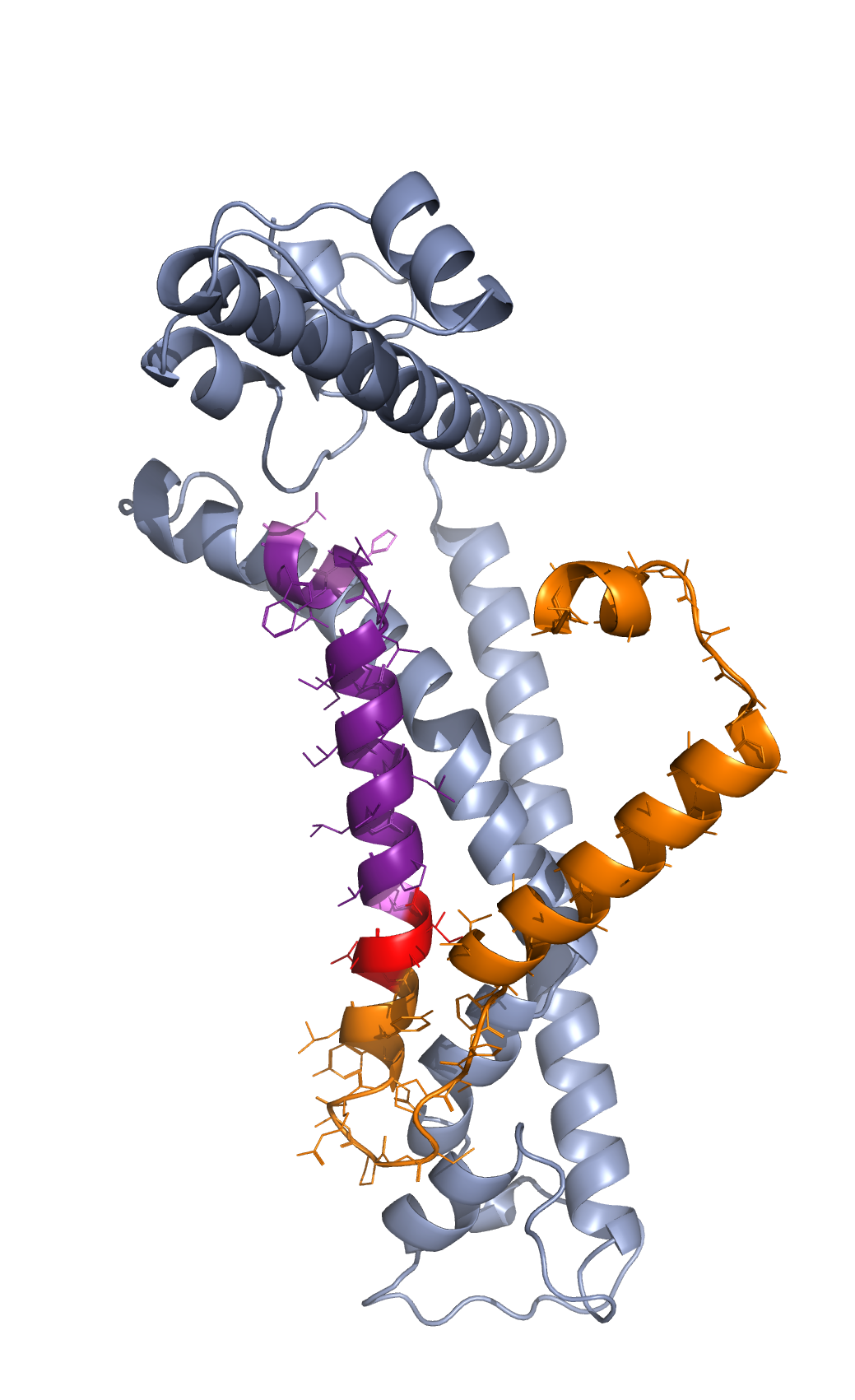
Caption: Coloring different segments of protein 6t0b chain m.#
Attributes#
Attribute |
Description |
|---|---|
|
name of a protein in the prefix of a PDB file name (e.g., 1xqf in 1xqfA.pdb) |
|
chain of a protein in the prefix of a PDB file name (e.g., A in 1xqfA.pdb) |
|
path where a target PDB file is place |
|
color of the entire protein |
|
names of segments |
|
which segments |
|
colors selected for the segments |
|
representation |
See also
Please see here for better understanding the file-naming system.
Output#
Finally, you will see the following output.
PyMOL(TM) Molecular Graphics System, Version 2.5.0.
Copyright (c) Schrodinger, LLC.
All Rights Reserved.
Created by Warren L. DeLano, Ph.D.
PyMOL is user-supported open-source software. Although some versions
are freely available, PyMOL is not in the public domain.
If PyMOL is helpful in your work or study, then please volunteer
support for our ongoing efforts to create open and affordable scientific
software by purchasing a PyMOL Maintenance and/or Support subscription.
More information can be found at "http://www.pymol.org".
Enter "help" for a list of commands.
Enter "help " for information on a specific command.
Hit ESC anytime to toggle between text and graphics.
Detected OpenGL version 4.6. Shaders available.
Detected GLSL version 4.60.
OpenGL graphics engine:
GL_VENDOR: NVIDIA Corporation
GL_RENDERER: NVIDIA GeForce RTX 2070/PCIe/SSE2
GL_VERSION: 4.6.0 NVIDIA 536.40
Detected 12 CPU cores. Enabled multithreaded rendering.
D:\Programming\anaconda3\envs\tmkit\Lib\site-packages\pymol\commanding.py:321: DeprecationWarning: setDaemon() is deprecated, set the daemon attribute instead
t.setDaemon(1)
