Concept#
TMKit contains seqNetRR, a high-performance computing library for constructing residue connections in sequence and contact-map context and performing the assignment of co-evolutionary features (or other types of co-variant features) in linear time. This module is aimed at facilitating co-evolutionary feature engineering, scheme design, and organization for machine learning modelling of interactions/contacts between residues.
In this tutorial, we will go through the definition of local residue-residue connections (LocRRCs) and the computational scheme to construct them at a fast speed.
LocRRCs. Connections between a residue of interest and its serially ordered neighboring residues are referred to as local residue–residue connections. It focuses on how one residue of a residue pair of interest connects with its neighbouring residues in the same side. The overall property of the unigraph of this residue is imparted by strengths between the edges in the graph. The strengths, in our case, are co-evolutionary features.
Assigning features to co-variant residue pairs in transmembrane proteins is particularly challenging due to their long sequences, which generate a large number of residue pairs.
The seqNetRR library significantly reduces computational time by implementing modular methods (minimize the overhead of frequent Python function calls) and utilising hash tables (efficient data retrieval and assignment).
Comparative analyses against Pandas and NumPy confirm that seqNetRR is the fastest method for assigning co-evolutionary features to LocRRCs, making it a highly optimised solution for large-scale transmembrane protein analysis.
Problem analysis#
Suppose we have a protein sequence of length L. The total number of residue pairs N extracted from the sequence is given by
All residue pairs can be represented as a 2D array or a 2D data object in Python. When a sliding window of size w is applied, the structure expands into a 3D data object, as illustrated in the plot below.
Constrained by t edges in a unipartite graph, the number of window-placed LocRRCs Nt for each residue is calculated as
leading to a total of 2×Nt LocRRCs for each residue pair.
The core challenge is efficiently assigning features to this large volume of residue pairs. A naive approach would result in an O(n³) time complexity, making it computationally infeasible for large datasets. However, by leveraging hashing techniques and optimizing the process to eliminate unnecessary computations, we can dramatically reduce runtime. Among various methods tested, the Hash-based approach has proven to be the most efficient.
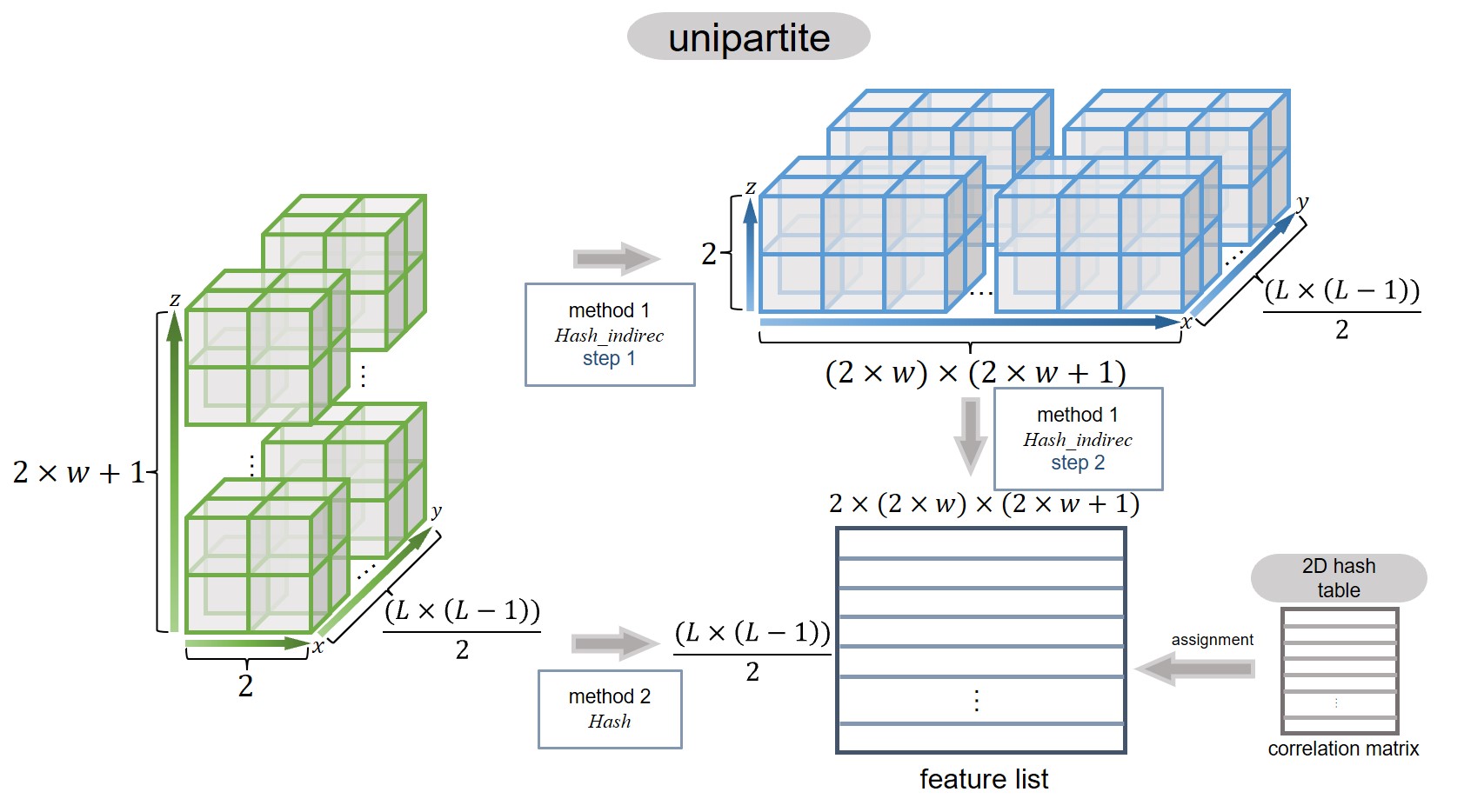
Caption: Computing scheme of assigning features to LocRRCs in seqNetRR.#
Method |
Description |
|---|---|
|
achieved with method 2 by assigning features and in the meantime constructing the entire list of residue pairs. Feature assignment is done by querying values from a hash table. |
|
achieved with method 1 by constructing the entire combinations of residues first and then going to feature assignment. |
|
achieved with method 2 by assigning features and in the meantime constructing the entire list of residue pairs. Feature assignment is done by pandas.at. |
|
achieved with method 2 by assigning features and in the meantime constructing the entire list of residue pairs. Feature assignment is done by numpy array. |
Runtime evaluation#
Tip
TRAIN, PREVIOUS, and TEST used in DeepHelicon[1] are used as benchmark datasets to gauge the runtime and downloaded via this link
Computing performance is gauged by the running time of different methods. We examined feature assignment based on LocRRCs constrained by a 5-node unipartite graph (refer to here). Given its acceptable CPU efficiency in handling LocRRCs, we included a comparison with the NumPy method. Notably, feature assignment using the Hash method has become a trivial task, with an average runtime of just 1.63s (1.64s for TRAIN, 1.94s for PREVIOUS, and 1.30s for TEST). The Hash method outperforms existing approaches, running 33 times faster than NumPy array indexing and 11 times faster than the Pandas pandas.at method, demonstrating superior efficiency in feature assignment for LocRRCs.
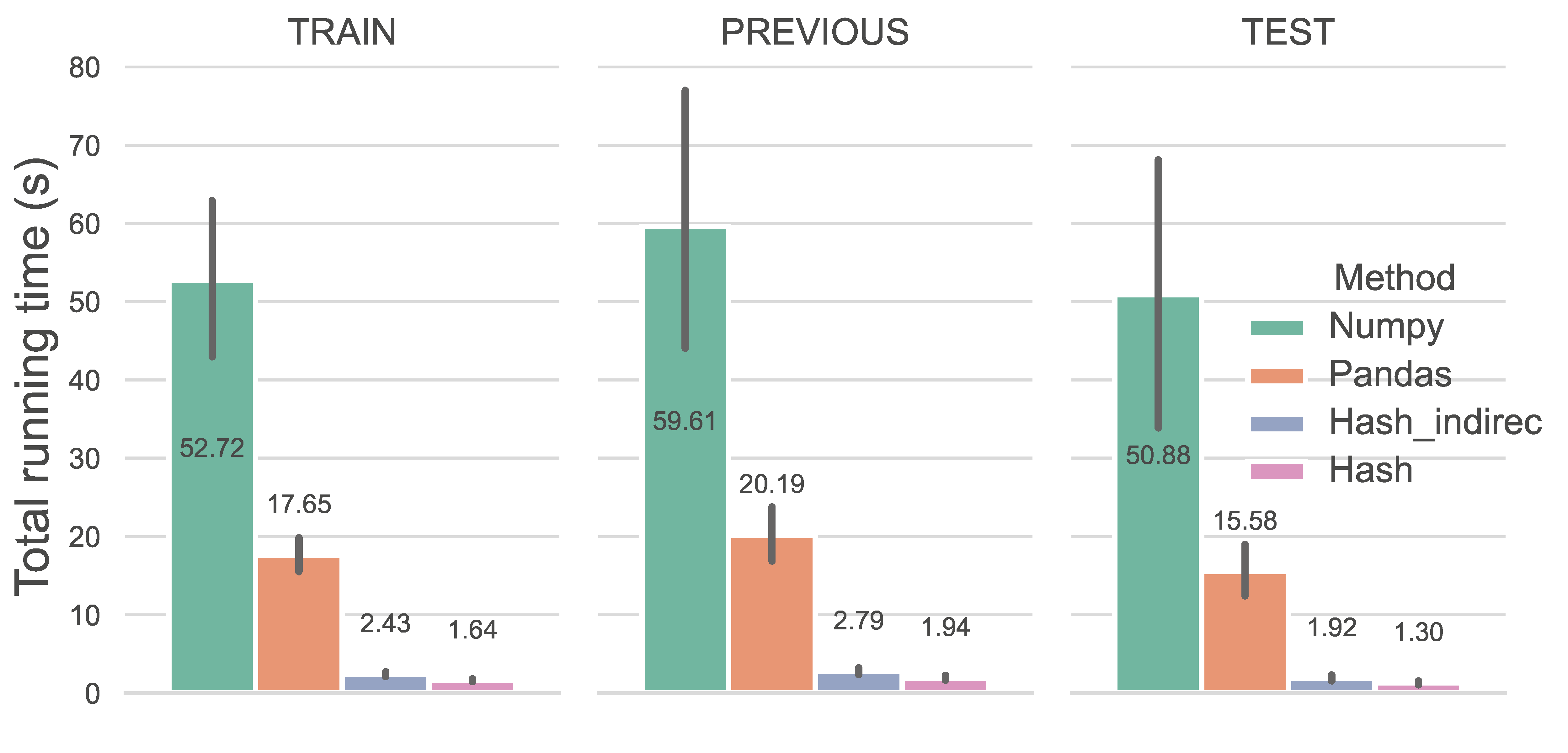
Caption: The runtime of the two tasks at the per-protein level.#
