LIPS program#
Introduction to LIPid-facing Surface#
TMKit integrates the LIPS (LIPid-facing Surface) method[1] to generate helix surfaces, LIPS scores, and entropy scores, facilitating the analysis of transmembrane protein interactions.
You can also refer to our recent publication[2] for further illustration. Transmembrane (TM) proteins are anchored to the membrane via α-helices, allowing them to interact with lipids. Therefore, studying lipid-accessible protein properties, such as helix orientation relative to lipids, is crucial. Below, we detail a helix orientation prediction process used in the LIPS method, which highlights the relationship between specialized structural features and the biological functions of membrane proteins.
Notably, TM proteins are enriched with coiled coils in TM regions, which contain heptad repeats—periodically recurring seven-amino-acid sequences labeled ABCDEFG. A heptad repeat generates seven distinct helical faces, with each of the seven residues alternately considered as an anchoring residue. According to Adamian and Liang[1], each anchoring residue is complemented by two adjacent residues (two positions apart), forming one of the seven surfaces. Thus, starting from the first residue A, the seven helical faces are ADE, BEF, CFG, DGA, EAB, FBC, and GCD, as demonstrated below.
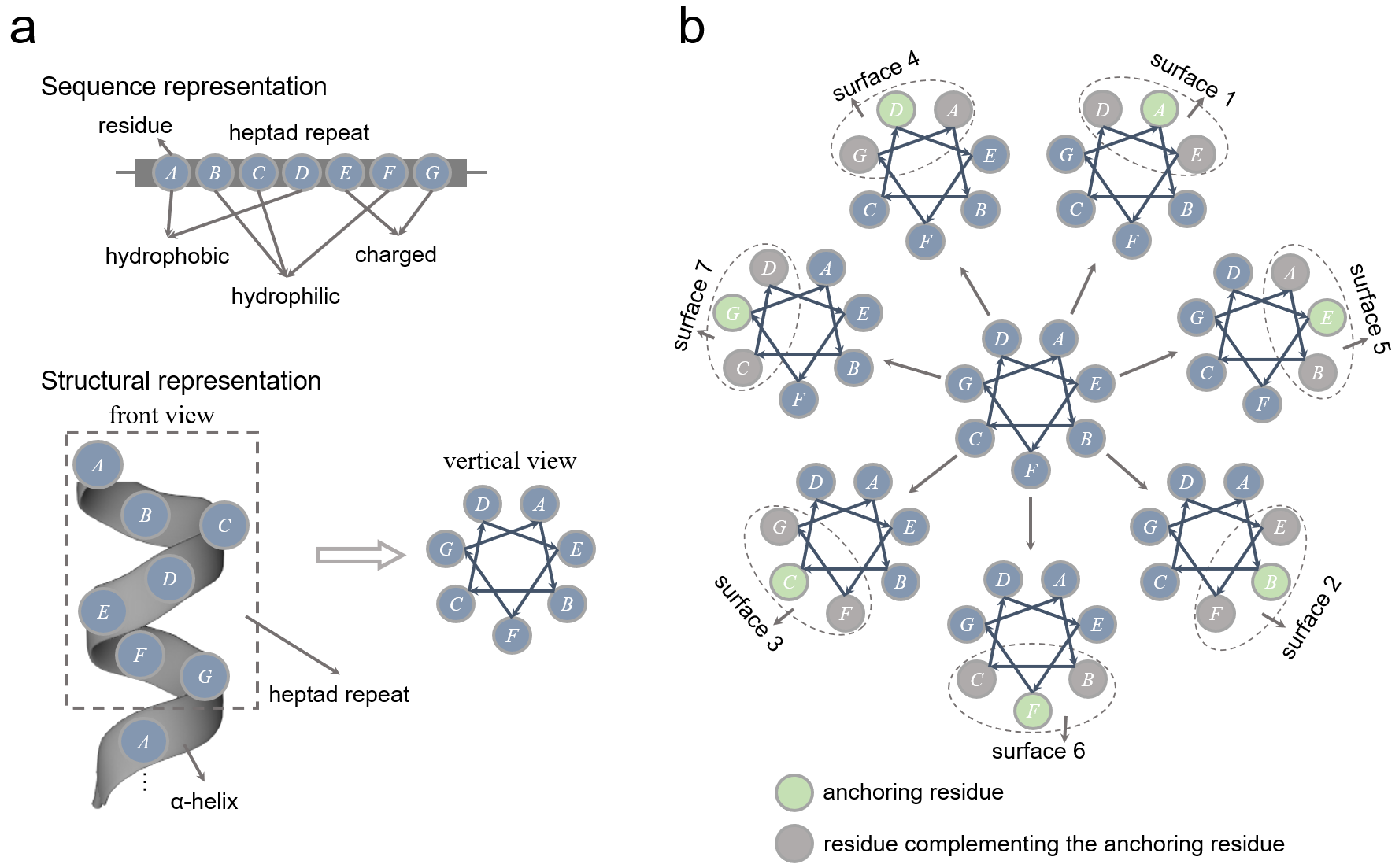
Caption: Schematic illustration of helical surfaces generated using the LIPS method. (a) shows the representations of the heptad repeat (seven residues ABCDEFG) in sequence and structural contexts. (b) shows the seven canonical surfaces of transmembrane α-helices, which are generated by taking each of the seven residues as an anchoring residue. Each surface consists of an anchoring residue and two residues complementing the anchoring residue. The seven surfaces are ADE, BEF, CFG, DGA, EAB, FBC, and GCD.#
In the LIPS pipeline, this helical partitioning is applied systematically:
Sliding through the TM protein sequence, each residue is assigned to one of the seven helical faces. Each helical face receives an entropy score and a lipophilicity score. These scores are integrated to compute the LIPS score, which estimates helix orientation. Since the majority of residues involved in TM helix interactions align with heptad repeats, both face-level LIPS scores and residue-level lipophilicity scores may aid in identifying interaction sites within TM proteins. In MBPred, the significance of helical face-related scores in predicting interaction sites is demonstrated through:
Mean Decrease in Impurity (Gini Importance), and Leave-One-Out Cross-Validation tests. These findings reinforce the functional relevance of heptad repeat-derived structural features in TM protein interactions.
Reminder of data
Please make sure that the build-in example dataset has been downloaded before you walk through the tutorial.
Example usage#
LIPS replies on an input of a multiple sequence alignment (MSA). We use the MSA of protein 1xqf chain A to generate LIPS results. We have put it in ./data/msa/.
Please note that TMKit includes a method that serves as a wrapper for the external lips.pl library, which can be accessed via the provided link. The lips.pl version in TMKit has been slightly modified from the original implementation, making it more convenient to retrieve results. The results will be saved in ./data/lips/.
See also
If you use this script or incorporate LIPS results in your studies, please also cite the original research[1].
Added in version 0.0.3: lips.pl was supported by this version of TMKit or above.
import tmkit as tmk
tmk.feature.generate_helix_surfaces(
msa_path='./data/msa/',
prot_name='1xqf',
file_chain='A',
sv_fp='./data/lips/',
)
If you have multiple proteins and want to get the LIPS results all at once, you can do it as follows. First, let’s generate a Pandas dataframe to describe the proteins.
import pandas as pd
prots = [
['1xqf', 'A'],
['3pux', 'G'],
['3rko', 'A'],
]
df_prot = pd.DataFrame(prots, columns=['prot', 'chain'])
Then, please put the dataframe to df_prot in command tmk.feature.bgenerate_helix_surfaces below. Please find the output and parameter illustration below.
import tmkit as tmk
tmk.feature.bgenerate_helix_surfaces(
msa_path='./data/msa/',
sv_fp='./data/lips/',
df_prot=df_prot,
)
Attributes#
Arribute |
Description |
|---|---|
|
name of a protein in the prefix of a PDB file name (e.g., 1xqf in 1xqfA.pdb) |
|
chain of a protein in the prefix of a PDB file name (e.g., A in 1xqfA.pdb) |
|
Pandas dataframe storing protein names and chain names |
|
path where a protein MSA file is placed |
|
path to save files |
See also
Please see here for better understanding the file-naming system.
Output#
We show below that for a single protein 1xqf chain A, what kind of output the program will give users. First, it has 7 files for 7 surfaces, each having their residues residing. Finally, it outputs a summary file with 7 surfaces with LIPOPHILICITY, ENTROPY, and LIPS scores.
save path is: tmkit/data/example/1xqfA/
SURFACE 0:
1 A 0.018 1.125
4 D 0.740 5.710
5 K 0.804 6.798
...
362 P 0.615 4.539
SURFACE 1:
2 V 0.026 1.158
5 K 0.804 6.798
6 A 0.573 4.896
...
362 P 0.615 4.539
SURFACE 2:
3 A 0.552 4.238
6 A 0.573 4.896
7 D 0.865 2.885
...
360 R 1.174 1.749
SURFACE 3:
4 D 0.740 5.710
7 D 0.865 2.885
8 N 0.679 5.217
...
361 V 0.751 3.144
SURFACE 4:
5 K 0.804 6.798
8 N 0.679 5.217
9 A 0.697 4.852
...
362 P 0.615 4.539
1 A 0.018 1.125
2 V 0.026 1.158
SURFACE 5:
6 A 0.573 4.896
9 A 0.697 4.852
10 F 1.621 2.735
...
360 R 1.174 1.749
2 V 0.026 1.158
3 A 0.552 4.238
SURFACE 6:
7 D 0.865 2.885
10 F 1.621 2.735
11 M 0.975 4.820
...
361 V 0.751 3.144
3 A 0.552 4.238
4 D 0.740 5.710
SURFACE LIPOPHILICITY ENTROPY LIPS
5 1.834 4.846 8.889
3 1.729 4.852 8.389
0 1.770 4.912 8.694
6 1.777 4.746 8.435
2 1.791 4.749 8.507
1 1.815 4.885 8.865
4 1.767 4.948 8.741
