PPI interface#
Identification of protein-protein interaction (PPI) interfaces of proteins is critical to understand the biological processes governed by them. Direct visualization of the PPI interfaces on 3D structures can facilitate their localization at the atomic coordinate level. TMKit is an open-source toolkit that enables access to the PPI interfaces by taking as input the structure of a protein of interest (POI) and a list of probabilities of residue sites to be involved in interactions. The program can automatically generate the label- or propensity-based PPI interfaces in between a POI and its interacting proteins (or its larger complex), which can be visualised in PyMOL.
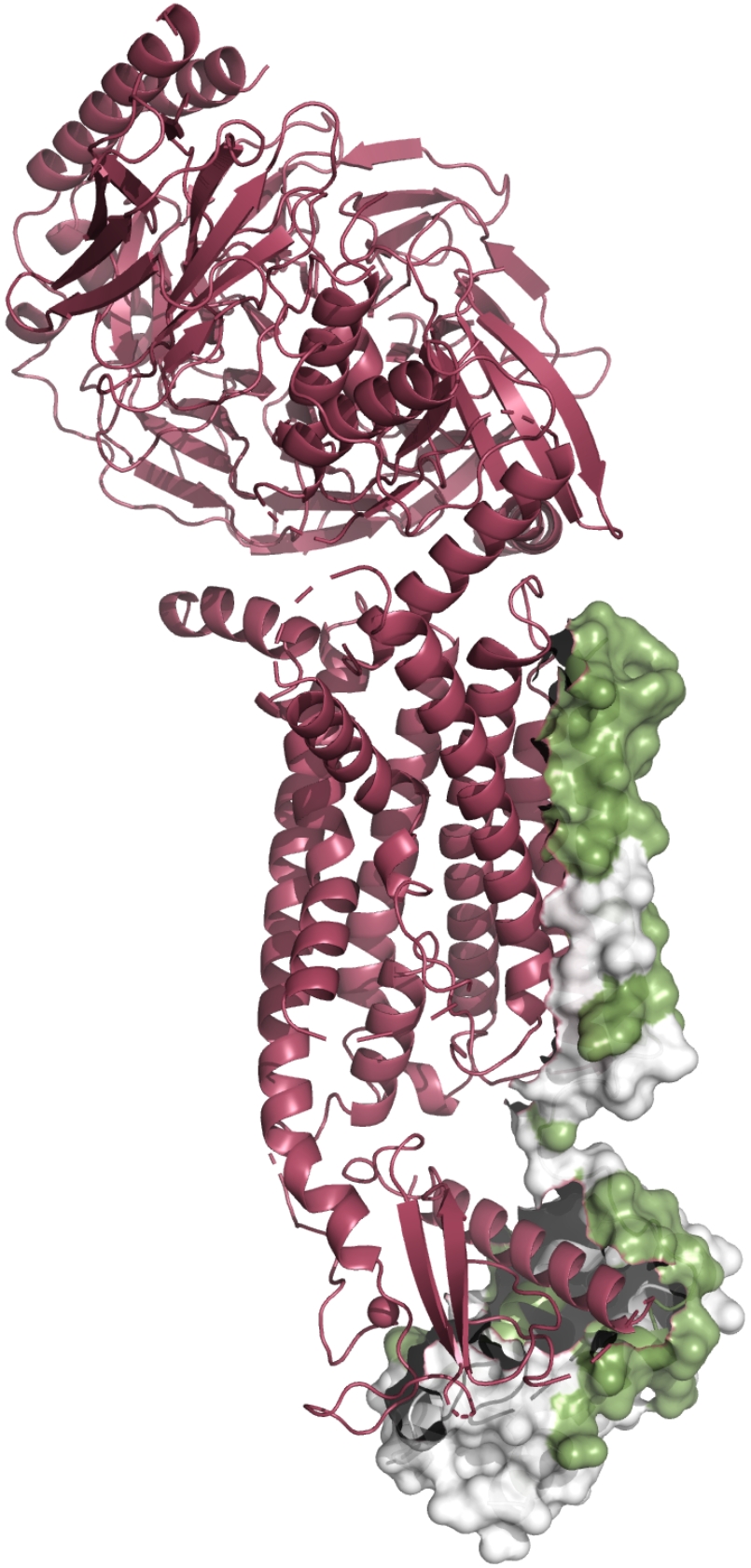
TMKit allows users to visualize the structure-based interfaces between proteins in a complex. As shown in the plot below, we generate the PPI interfaces in this paper[1].
Caption: Interfaces of 5b0w chain A and 6t0b chain m in complex with their interaction partners, respectively.#
Attention
Visualization of PPI interfaces in TMKit relies on PyMOL but it is not installed by default. To use this function, you need to install PyMOL. There are two ways to install it very easily as below. We recommend installing it using an open-source PyMOL version (pymol-open-source). For more detail, see also here.
# recommended option
conda install -c conda-forge pymol-open-source
# or
conda install -c schrodinger pymol
Tip
Please make sure that the build-in example dataset has been downloaded before you walk through the tutorial.
Example usage#
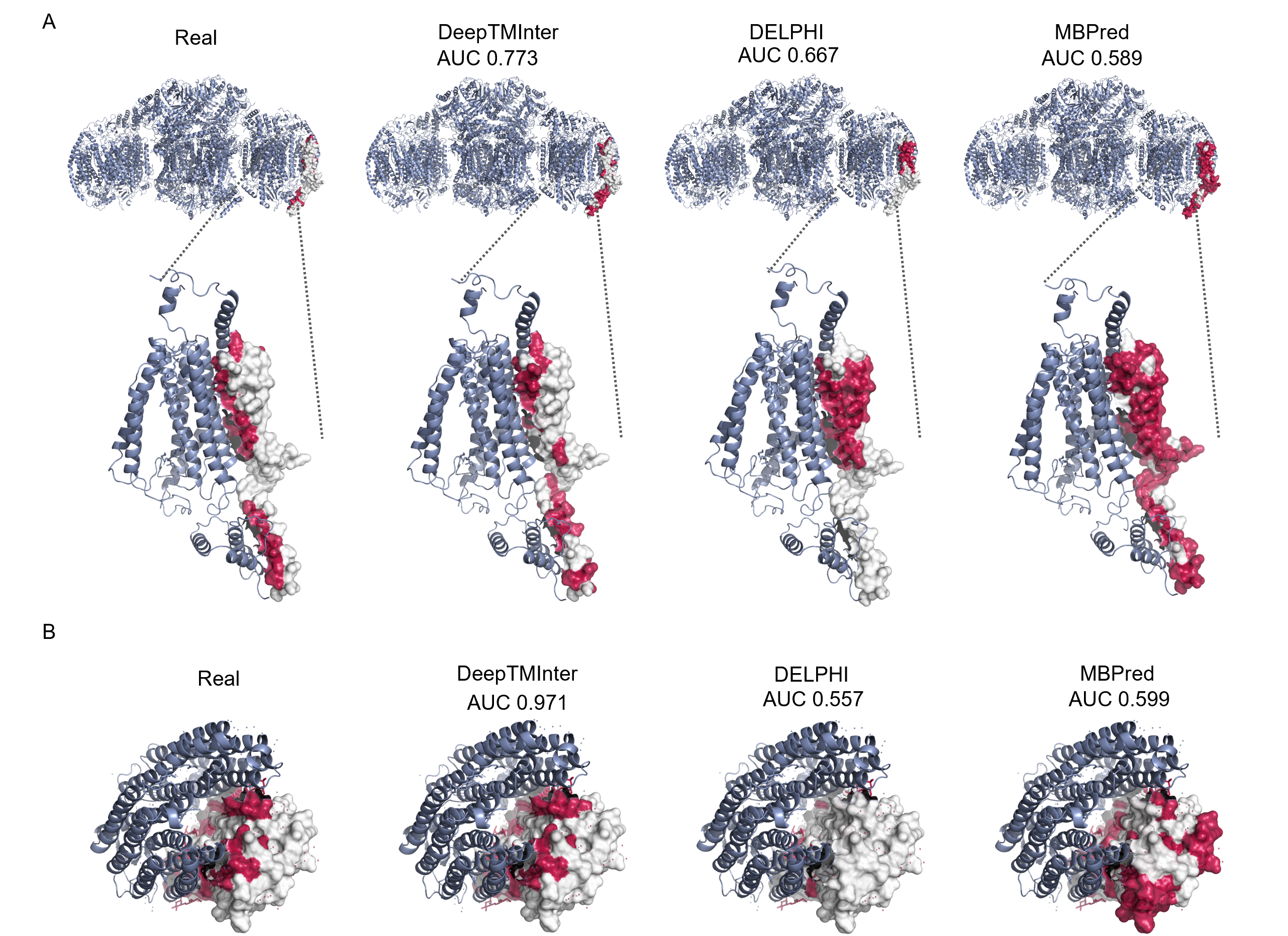
After importing TMKit, the following command allows you to visualize the PPI interfaces of protein 6e3y chain E with other chains in complex protein 6e3y. The PPI interfaces are predicted using DeepTMInter[2].
import tmkit as tmk
tmk.vs.protoc_deeptminter(
prot_name='6e3y',
prot_chain='E',
pdb_chain_fp='data/pdb/',
dist_fp='data/rrc/',
pdb_complex_fp='data/pdb/pdbtm/',
tool='deeptminter',
draw_type='probability',
isite_fp='data/isite/',
sv_bfactor_fp='data/vs/bfactor/6e3y/',
pymol_bg_chain_ids='B+C+J+K',
)
Caption: Predicted PPI interfaces in green for 6e3y chain E in complex with their interaction partners in red, respectively.#
The keynote here is the input of predicted PPI interfaces. For your information, we show what this input file looks like below.
Residue ID |
Residue |
Predicted probability |
|---|---|---|
1.0 |
E |
0.18815077418058843 |
2.0 |
A |
0.18221259814512925 |
3.0 |
N |
0.48554193714932137 |
… |
… |
… |
114.0 |
K |
0.7522871745416599 |
114.0 |
R |
0.7660381644357218 |
In fact, you can generate a file in this format if you have other predicted PPI interfaces by parameter isite_fp. Then, TMKit will recognize that anyway.
It should also be noticed that TMKit will use the Predicted PPI interfaces to generate a file that is recognized by PyMOL as saved in sv_bfactor_fp.
Attributes#
Attribute |
Description |
|---|---|
|
name of a protein in the prefix of a PDB file name (e.g., 1xqf in 1xqfA.pdb) |
|
chain of a protein in the prefix of a PDB file name (e.g., A in 1xqfA.pdb) |
|
path where a target PDB file is place |
|
path where a file containing actual residue distances (e.g. ./data/rrc in the example dataset) |
|
path where a PDB file showing a protein complex is placed |
|
tool name. Currently, the reading of DeepTMInter, DELPHI, and MBPred files is supported |
|
path where a file showing interaction sites and the interaction likelihoods is placed |
|
path to save a bfactor file |
|
interaction chains in a protein complex |
|
color of interaction chains in a protein complex |
|
label_actual, # label_actual, label_predicted, probability |
See also
Please see here for better understanding the file-naming system.
Output#
Finally, you will see the following output.
PyMOL(TM) Molecular Graphics System, Version 2.5.0.
Copyright (c) Schrodinger, LLC.
All Rights Reserved.
Created by Warren L. DeLano, Ph.D.
PyMOL is user-supported open-source software. Although some versions
are freely available, PyMOL is not in the public domain.
If PyMOL is helpful in your work or study, then please volunteer
support for our ongoing efforts to create open and affordable scientific
software by purchasing a PyMOL Maintenance and/or Support subscription.
More information can be found at "http://www.pymol.org".
Enter "help" for a list of commands.
Enter "help command-name" for information on a specific command.
Hit ESC anytime to toggle between text and graphics.
======>Time to read&label distances for 6e3y E: 0.004998445510864258s.
======>Time to read&label distances for 6e3y E: 0.003000020980834961s.
['P', 'N', 'A', 'B', 'G', 'R', 'E']
bFactor:
///E/29/N new: 0.188151
///E/29/CA new: 0.188151
///E/29/C new: 0.188151
...
///E/143/NH2 new: 0.766038
Detected OpenGL version 4.6. Shaders available.
Detected GLSL version 4.60.
OpenGL graphics engine:
GL_VENDOR: NVIDIA Corporation
GL_RENDERER: NVIDIA GeForce RTX 2070/PCIe/SSE2
GL_VERSION: 4.6.0 NVIDIA 536.40
Detected 12 CPU cores. Enabled multithreaded rendering.
D:\Programming\anaconda3\envs\tmkit\Lib\site-packages\pymol\commanding.py:321: DeprecationWarning: setDaemon() is deprecated, set the daemon attribute instead
t.setDaemon(1)
